What Other Tales Can Coral Skeletons Tell?
Scientists strive to get into the genes of fossil corals to extract their evolutionary history
In 2003, we traveled by ship to the New England Seamounts—a chain of extinct, undersea volcanoes about 500 miles off the East Coast of North America—to help collect dead corals that have been on the bottom of the ocean for thousands of years. For scientists, these corals are like deep-sea time machines.
As they grew, the skeletons of these long-living corals incorporated trace chemicals from surrounding seawater. By analyzing chemical variations in the corals’ annual growth bands, our colleagues, geochemist Jess Adkins of the California Institute of Technology and Laura Robinson of Woods Hole Oceanographic Institution, are reconstructing the history of how ocean and climate conditions have changed over Earth’s history. (See “The Coral-Climate Connection.”)
But as we pulled corals from the sea, we began to wonder if we could go one step further and unlock the genetic makeup of these fossil coral species. Perhaps they could tell us the story of how deep-sea coral populations evolved over thousands of years—in response to the ocean and climate shifts that their skeletons are recording. Accomplishing this, however, means achieving something never done before: extracting DNA from these long-dead organisms.
A new line of inquiry
Tapping into the corals’ ancient genetic record could let us see how populations adapted to changes in their environment over time. We would be able to determine if, and when, they moved among different seamounts at specific periods in history. We could also learn if populations preferred living at one depth or another at different times in history.
By matching genetic information with the dates of climate events recorded in their skeletons, we would determine whether populations migrated away from particular seamounts at times when the climate and ocean warmed or cooled, or if the populations were vastly reduced to a few survivors. All of this would give us the first insights into how corals were reacting to climate changes as far back as 100,000 years ago.
Unlocking genetic data from coral skeletons opens up a completely new avenue of coral and climate research. If successful, it would be possible to use these methods to extract genetic data from other dead coral populations worldwide. These techniques could be used, for example, to advance understanding of shallow-water coral reefs that died because of periodic ocean water warming associated with El Niño events.
It could also be possible to apply these techniques to other fossilized marine species, including clams, mussels, and snails from hydrothermal vents, to determine how they evolved through time. (See “The Evolutionary Puzzle of Seafloor Life.”)
Coral collecting on the seafloor
The corals we study, called scleractinians or stony corals, have been living for about 250 million years throughout the world’s oceans, from polar to tropical regions. We’re interested in two species, Desmophyllum dianthus and Lophelia pertusa, which survive in sunless areas of the ocean up to 4,000 meters (2.5 miles) deep by filtering krill, copepods, and other tiny food particles drifting through the water column.
During three expeditions—one in 2003 with Adkins, and two more in 2005—we gathered several thousand dead scleractinian corals from 15 different seamounts. On two expeditions, we collected corals using the WHOI-operated submersible Alvin; on one, we used the remotely operated vehicle Hercules, operated by University of Rhode Island.
Operators of both vehicles gathered the samples by placing nets in the vehicles’ robotic claws and sweeping them over the seafloor. They collected hundreds of fossil corals, ranging in size from tennis balls to volleyballs, in each sweep of the net. In other areas of the seafloor, dozens of dead corals were stuck together on overhangs, so the vehicles could simply snap off large pieces and bring them back to the surface.
Onboard the research vessels, we made detailed notes about the corals’ appearances, took photographs, and then sorted, cleaned, and froze them. We also assembled digital data—maps, photo, and video documentation, and information about environmental conditions in which we found them, such as temperature, salinity, and current speed.
Avoiding contamination
Sensitive methods are required for analysis in long-dead coral samples, because DNA doesn’t stick around forever. Unless samples are kept at a constant cold temperature, or are preserved properly, DNA degrades very quickly over time.
Fortunately, the corals we study lived and died in deep, cold water—and were protected by sediments—so we theorized that there would be a high likelihood that DNA remained within the skeleton. We were right.
Still, challenges remain. Familiar methods used routinely to extract genetic information are not sensitive enough for the small amounts of DNA left in dead coral skeletons. Also, not all the corals we collected will still contain DNA; some will have degraded. So we have to try a variety of methods to determine what works best. Even the most accomplished ancient DNA studies have only a 25 percent success rate with their samples.
There are few marine population studies using ancient DNA, and this project is among the first studies of ancient marine invertebrates. However, we are not the first scientists to extract ancient DNA from long-dead organisms. Scientists have extracted DNA from extinct land-dwelling animals, such as bison, woolly mammoths, and musk ox, some dating back 30,000 years. Researchers have also revealed how genetic populations have changed through time for species of trees, shrubs, mosses, and herbs, some as old as 400,000 years.
Work by these scientists has shown us that the DNA extraction methods must be exact, because the risk of contamination is huge. A random strand of hair or a stray speck of skin could throw off the results. Even paper is not allowed in ancient DNA laboratories, because the DNA contained in paper—previously living trees—could ruin samples undergoing analyses.
Trial and error
In April 2006, Rhian spent two weeks at the Paleo-DNA Laboratory at Lakehead University in Thunder Bay, Canada, which specializes in pinpointing methods for this type of specialized work. They had never done work on corals, so they were excited to help us refine our methods. Getting the optimal mix of chemicals and processes has been the result of time-consuming trial and error. All the testing has been worth it, though. We now have ever-improving methods to extract DNA and can concentrate on the project’s larger goals.
The genetic data from the corals will be combined with Adkins’ coral age and climate data to make correlations between the two. Then we will draw conclusions about what happened deep in the oceans, during past climate changes, and even make predictions about the future, including what will happen to deep-water corals (and the many animals that live on them) if the climate changes.
This project was funded by the WHOI Ocean Life and Ocean Climate Change Institutes. Research cruises were supported by the National Science Foundation and the NOAA Ocean Exploration Program.
Slideshow

Slideshow
 A thriving scleractinian coral community on Nashville Seamount, located on the New England Seamounts. A Desmophyllum dianthus coral (with orange tentacles sticking out) has a large brittle star crawling on top. The coral Lophelia pertusa is shown at the lower right corner, along with many orange octocorals. (Photo courtesy of DASS05_IFE_IAO_URI_NOAA)
A thriving scleractinian coral community on Nashville Seamount, located on the New England Seamounts. A Desmophyllum dianthus coral (with orange tentacles sticking out) has a large brittle star crawling on top. The coral Lophelia pertusa is shown at the lower right corner, along with many orange octocorals. (Photo courtesy of DASS05_IFE_IAO_URI_NOAA)- Scientists gathered dead coral samples by placing nets in the robotic claws of deep-sea vehicles. They collected hundreds of corals, ranging in size from tennis balls to volleyballs, in each sweep of the net. (Photo courtesy of DASS05_IFE_IAO_URI_NOAA)
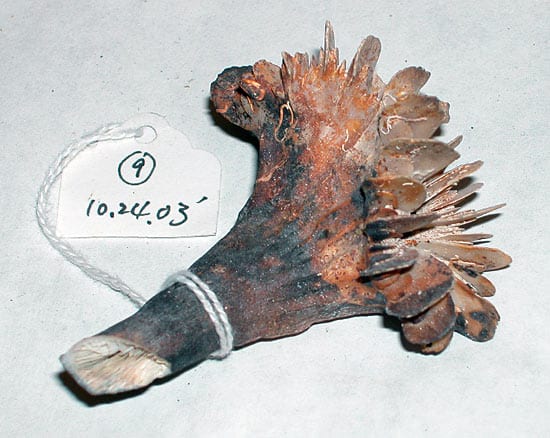
- One of the thousands of Desmophyllum dianthus coral samples collected and catalogued for future climate and DNA studies. (Photo courtesy of Rhian Waller, Woods Hole Oceanographic Institution)
- To avoid contaminating coral samples, WHOI scientist Rhian Waller wore a protective suit, mask, and gloves while working at the Paleo-DNA Laboratory at Lakehead University. (Photo courtesy of Rhian Waller, Woods Hole Oceanographic Institution)
Video
Related Articles
Featured Researchers
See Also
- Tim Shank, Research Lab at WHOI
- Rhian Waller's Home Page
- The Coral-Climate Connection from Oceanus magazine
- Paleo DNA lab at Lakehead University
- Dive and Discover: The New England Seamounts
- Dive and Discover: Interview with Rhian Waller
- Dive and Discover: Interview with Tim Shank
- Who is Jess Adkins?
- The Evolutionary Puzzle of Seafloor Life Oceanus magazine article by Tim Shank
- More about Tim Shank Includes video interviews, biography, and career timeline
- Coral Gardens in the Dark Depths from Oceanus magazine
- NOAA Ocean Explorer: 2005 expedition to collect North Atlantic corals
- Lullabye for Larvae from Oceanus magazine