Revealing the Ocean’s Invisible Abundance
Scientists develop new instruments to study microbes at the center of the ocean food web
Microbes. They are invisible to the naked eye, but they play a critical role in keeping our planet habitable. They are everywhere, in abundant numbers, but are still difficult to find. They come in a multitude of varieties, but too often are difficult to distinguish from one another.
Wherever there is water (fresh or salt), there are usually microbes—microscopic, single-celled organisms. In the ocean, they form an unseen cornucopia at the center of a food web that ultimately nourishes larger organisms, fish, and people.
Their fundamental role in the ocean’s food supply makes them critical targets for study, and scientists would like to know much more about them. They would like to identify them and count them. They would like to learn more about how marine microorganisms (part of what we call plankton) eat, grow, reproduce, and interact with other organisms. They would like to determine how changes in the ocean might affect the microbial communities’ vitality and viability.
Finding minuscule life forms in a seemingly infinite ocean isn’t trivial. But in recent years, oceanographers have been developing new techniques and instruments to identify and count marine microorganisms. Year by year, we are learning more and more about them and discovering that they are even more numerous, varied, and important than we previously thought.
A diverse microbial community
Some marine microbes are bacteria, or prokaryotes—simple cells with no specialized organelles, which are among the smallest of living things. Others are eukaryotes—larger and more complex cells with a nucleus, mitochondria, and other organelles.
Eukaryotic microbes, also called protists, include both producers, such as algae, and consumers, such as protozoa. They thrive in a variety of habitats—living suspended in the water, in bottom sediments, or on other objects. They form communities, or assemblages, of different species that photosynthesize, consume each other, and are, in turn, consumed by other things in the ocean’s food web.
In the last few years, we have considerably advanced our knowledge of the structure and function of these assemblages—particularly planktonic assemblages that we sample by collecting the water they inhabit. We now know that these plankton assemblages are diverse, composed of species with widely different sizes, growth rates, and nutrition. Not surprisingly, we know more about the larger protists (greater than 100 microns) than the smaller ones (under 20 microns). Larger protists are easily visible using light or electron microscopes. They have features that remain intact throughout procedures to sample, preserve, and examine them, which can break or distort cells. These features are often lacking in the smaller organisms; and if they are present, they are harder to see and characterize.
Identifying protists has always involved some type of microscopic analysis, with someone looking at the shapes, or morphology, of the cells. But now we also use molecular methods—techniques that give scientists the ability to detect and identify the presence of even small protists based upon their DNA in water samples. Scientists have begun to describe the genetic composition of communities of species that live and interact in the same water. Our next objective is to overcome several technical challenges so that we can routinely monitor changes in protist populations over time.
Sampling the invisible
So far, all of our detection and identification techniques, both morphologic and molecular, have relied on collecting samples from remote sites and analyzing them in laboratories. But these techniques don’t give us all the information we need.
Collecting samples from ships means physically taking separate water samples, at separate times, in separate places. Samples taken this way are, quite literally, just single samples—of one location at one time. They don’t provide a continuous picture of protists in a given area of the ocean. And they don’t allow us to detect how the protists respond to rapidly changing environmental conditions.
What researchers want is the ability to collect and analyze samples over long time periods in the ocean, to have a continuous sampling and recording procedure, and to obtain data in as close to real time as possible.
Overcoming engineering hurdles
Several technical challenges, however, still make it difficult to remotely detect and count microbes in their own environment. One is the number of organisms, or microscopic cells, in a given water sample. In most marine planktonic environments, microbes are present in low numbers and organisms targeted for study may only be a small proportion of the total population. To overcome this low density, researchers in the laboratory must often concentrate several liters of water into a much smaller volume for analysis by passing it through filters designed to retain the protists, then resuspending them in smaller volumes for analysis.
Once water samples are collected and concentrated, microbes can be analyzed in several ways, so automated systems must be designed to accommodate the analysis method. For instance, if scientists want to use only the organisms’ genetic material to identify them, collection systems must be able to break open cells and collect their DNA. If they want to study the whole organisms, though, the systems must keep the cells intact.
In fact, researchers are already developing instruments that can either detect a genetic signal from a microbial population or monitor one of its biological activities—and do it autonomously, without requiring scientists to be on the scene.
They can be pre-programmed to collect water samples over time periods ranging from hours to months and spaces ranging from inches to miles—depending on the particular microbes and biological activities the scientists want to study. These instruments inject water into flexible bags containing a solution that preserves the cells for later examination.
SID, ESP, and FlowCytobot
Three examples of instruments for remote analysis of marine microbes do solve many of the technical problems.
The Environmental Sample Processor, ESP, developed by Chris Scholin at Monterey Bay Aquarium Research Institute (MBARI), attaches to a mooring anchored to the ocean bottom and collects and preserves water samples. It extracts nucleic acids from the protists in the water and detects specific organisms by their DNA. It can also preserve samples for microscope analysis in the laboratory. Researchers have already used it to detect species that cause harmful algal blooms and to distinguish types of planktonic larvae in the ocean. It will soon have even greater capacity to detect and distinguish organisms.
The Submersible Incubation Device, SID, a moored instrument developed by Craig Taylor at WHOI, determines levels of photosynthesis in the water around it by robotically measuring carbon dioxide taken up by phytoplankton in the samples. Up to 50 of these experiments can be performed before the instrument needs to be removed from the ocean to analyze the samples and determine what species are present.
A third instrument, FlowCytobot, is a submersible flow cytometer— a device that counts single cells flowing through it. Developed by Robert Olson at WHOI, it is also anchored to the seafloor near the coast. It counts and analyzes microbial cells in the water continuously for up to two months. FlowCytobot identifies microbes by the way they scatter light, or by the way certain pigments in the cells emit fluorescent light. Because it samples continuously, scientists can see changes in plankton populations over time that cannot be detected by traditional sampling.
A coastal observatory network
The ultimate goal is a continuous, remote system that can detect, distinguish and count microbes in the environment. In the laboratory, scientists can do all these things by filtering samples, identifying DNA within them, and examining microbes under microscopes. But designing, programming, and building a system to carry out all of these steps remotely is a challenge.
One of the difficulties for this work is that DNA analysis requires heat, which requires power. Remotely deployed instruments depend on batteries for power, and adding batteries quickly makes instruments too heavy, big, and costly to build. To overcome this hurdle, scientists have sought a viable alternative; developing long-term installations of instruments powered by cables from a nearby shore.
In recent years, several coastal ocean observatories have been built that have cables linking power nodes on the ocean floor with shore-based facilities. One of these is near Woods Hole, at the Martha’s Vineyard Coastal Observatory (MVCO). Instruments plugged into seafloor nodes receive power from the cables and transmit data back via the cables.
This level of available power has stimulated the development of new biological sensors and methods that will let scientists take measurements continuously and accurately.
In the lab, we are working to develop and assemble several instrument modules into the FlowCytobot automated system to install at the MVCO. The system will detect microbial cells, identify them genetically, and obtain accurate counts of particular species. It will let us monitor specific microbial populations that play significant roles in the food web and detect changes taking place, on a daily basis.
The development of new sensors is also important to national efforts to build an infrastructure of ocean observation systems. Ocean observatories are the wave of the future in many fields of oceanography. Some will monitor coastal water; others will monitor the open ocean. Many already exist, and many more are being planned, through several national programs. These programs will incorporate existing coastal observatories into a network, expanding their research capabilities, and building more at key coastal sites. We will use the observatories, each with seafloor cables supplying power, to collect and share information on a previously intractable microbial world – the broad group of tiny cells that control the coastal ocean’s food supply.
A Quick Guide to Ocean Observation Systems Acronyms
The U.S. Network of Ocean Observation Systems will collect continuous, reliable information about our coastal ocean. Moored and mobile installations will carry instruments and sensors that sample and measure environmental variables, then transmit the data to computer systems that store, analyze, and model the data to describe and predict oceanic conditions. Some of the following acronyms denote permanent observatories in coastal water, some denote deep or open water observing systems, and some denote programs or organizations that have responsibility for planning or oversight of ocean-observing networks.
COOS Coastal Ocean Observing Systems The U.S. has more than forty observatory installations to monitor coastal ocean conditions, with more planned.
IOOS Integrated and Sustained Ocean Observing System The nationwide network of ocean observatories on platforms such as ships, airplanes, satellites, buoys, and drifters.
GOOS Global Ocean Observing System An international program to create a permanent global system for ocean observation.
ECCOO Eastern Consortium of Coastal Ocean Observatories Observatories on the U.S. East Coast
MVCO Martha’s Vineyard Coastal Observatory Operated by WHOI, and provides real time oceanographic and meteorological data
GoMOOS Gulf of Maine Ocean Observing System National pilot program posting hourly oceanographic data from the Gulf of Maine.
LEO Long-term Ecosystem Observatory Observatory operated by Rutgers University off the coast of New Jersey, collecting data from satellites, aircraft, ships, moorings, and autonomous underwater vehicles.
Slideshow

Slideshow
 SHORE TO SHIP—WHOI researchers Alexi Shalapyonok, Heidi Sosik, and Robert Olson (left to right) carefully load the FlowCytobot onto a WHOI research vessel for installation on the seafloor at the Martha’s Vineyard Coastal Observatory. The instrument counts and identifies protist cells in the water, and the data are transmitted via undersea cable back to shore. (Tom Kleindinst, WHOI Graphic Services.)
SHORE TO SHIP—WHOI researchers Alexi Shalapyonok, Heidi Sosik, and Robert Olson (left to right) carefully load the FlowCytobot onto a WHOI research vessel for installation on the seafloor at the Martha’s Vineyard Coastal Observatory. The instrument counts and identifies protist cells in the water, and the data are transmitted via undersea cable back to shore. (Tom Kleindinst, WHOI Graphic Services.) OVER THE SIDE—the Submersible Incubation Device (SID) hangs from a cable, ready to be moored on the sea bottom, where it will take samples of surrounding seawater and measure photosynthesis in the ocean. (Craig Taylor, WHOI.)
OVER THE SIDE—the Submersible Incubation Device (SID) hangs from a cable, ready to be moored on the sea bottom, where it will take samples of surrounding seawater and measure photosynthesis in the ocean. (Craig Taylor, WHOI.) TIRELESS UNDERSEA WORKER—The robotic Environmental Sample Processor (ESP) lifts off the deck and begins its journey to the seafloor off Monterey, California. It will be moored there for a lengthy stay and take repeated samples of protists in the water. (Kim Fulton-Bennett, ©2004 MBARI.)
TIRELESS UNDERSEA WORKER—The robotic Environmental Sample Processor (ESP) lifts off the deck and begins its journey to the seafloor off Monterey, California. It will be moored there for a lengthy stay and take repeated samples of protists in the water. (Kim Fulton-Bennett, ©2004 MBARI.)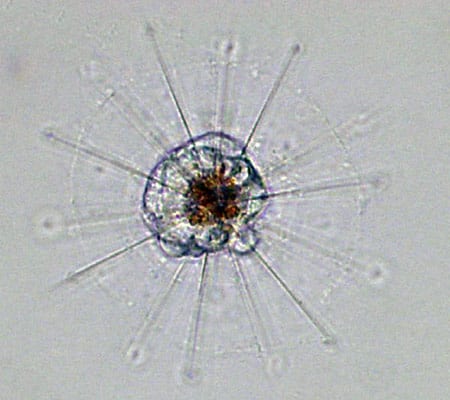 A heliozoan is heterotrophic, meaning it consumes plant and animal matter including even small animals. (Dawn Moran, WHOI.)
A heliozoan is heterotrophic, meaning it consumes plant and animal matter including even small animals. (Dawn Moran, WHOI.)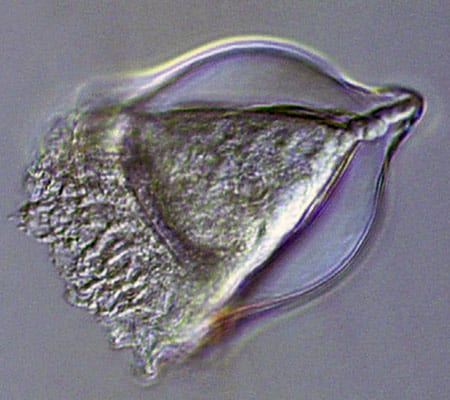 Tintinnids have transparent vase-like shells for protection, and are consumers of a wide variety of cells and detritus. (Dawn Moran, WHOI.)
Tintinnids have transparent vase-like shells for protection, and are consumers of a wide variety of cells and detritus. (Dawn Moran, WHOI.)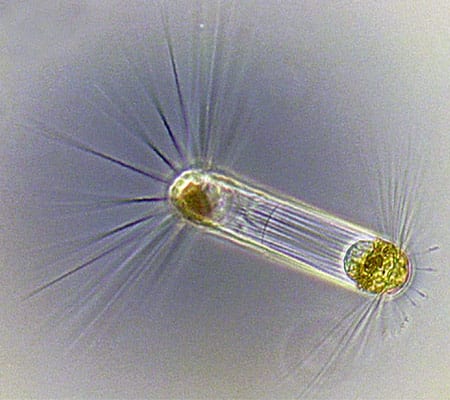 Diatoms (this one, Corethron) are at the base of the food chain, using photosynthesis to live, grow, and multiply. (Dawn Moran, WHOI.)
Diatoms (this one, Corethron) are at the base of the food chain, using photosynthesis to live, grow, and multiply. (Dawn Moran, WHOI.)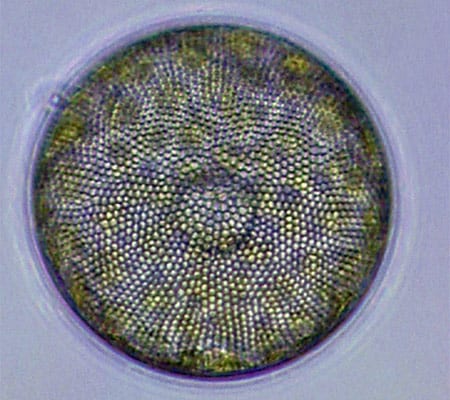 Viewed end-on, the diatom Coscinodiscus is a study in symmetry and pattern, reminiscent of a sunflower's seeds. (Dawn Moran, WHOI.)
Viewed end-on, the diatom Coscinodiscus is a study in symmetry and pattern, reminiscent of a sunflower's seeds. (Dawn Moran, WHOI.)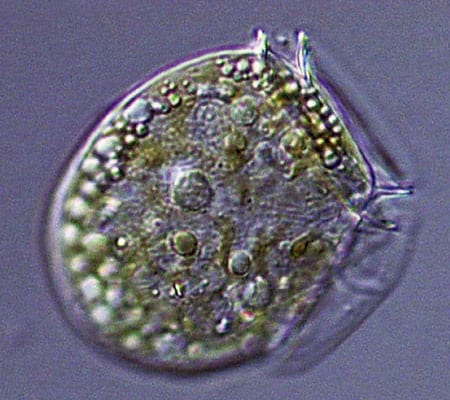 The dinoflagellate Dinophysis, plump and harmless-looking, produces a toxin that causes diarrhetic shellfish poisoning. (Dawn Moran, WHOI.)
The dinoflagellate Dinophysis, plump and harmless-looking, produces a toxin that causes diarrhetic shellfish poisoning. (Dawn Moran, WHOI.)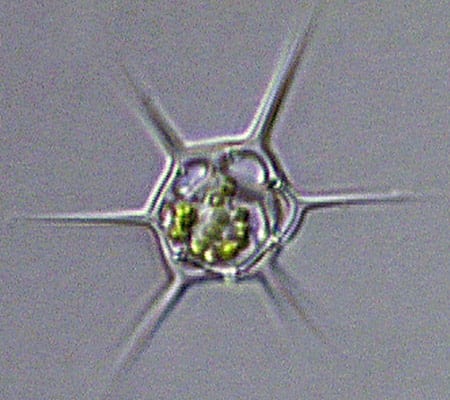 Dictyocha, a silicoflagellate, has an intricate, internal glassy skeleton and a starry shape that helps it avoid sinking. (Dawn Moran, WHOI.)
Dictyocha, a silicoflagellate, has an intricate, internal glassy skeleton and a starry shape that helps it avoid sinking. (Dawn Moran, WHOI.)
Related Articles
- Our eyes on the seafloor
- Bridge-to-PhD program at WHOI opens doors for new scientists
- A coral reef kickstart
- Junk Food
- A Lobster Trap for Microbes
- The Unseen World on Coral Reefs
- The Recipe for a Harmful Algal Bloom
- The Bacteria on Your Beaches
- Scientists Reveal Secrets of Whales
- In the Gardens of the Queen
- The Amazing Acquired Phototroph!
- What Happened to Deepwater Horizon Oil?
- New Device Reveals What Ocean Microbes Do
- Life Dwells Deep Within Earth’s Crust
- Can Animals Live Without Oxygen?