Marine Microbes vs. Cystic Fibrosis
Scientist seeks medicines from nature
Microbes that grow in the ocean could one day help doctors combat the deadly disease cystic fibrosis (CF), said Tracy Mincer, a microbiologist at Woods Hole Oceanographic Institution.
Mincer studies the small chemicals marine microbes make to do specific jobs such as communicating with each other or warding off predatory protozoa. In October 2011, the Flatley Discovery Lab (FDL) of Charlestown, Mass., awarded Mincer a $1.2-million grant to provide thousands of the chemicals for screening as potential drugs to treat CF.
The concept of using chemicals made by bacteria or fungi to treat diseases is hardly new. Penicillin came from the bread mold Penicillium and streptomycin came from bacteria known as actinomycetes, to name two well-known examples. But looking for medically valuable substances in the ocean is relatively new—and very promising. Each kind of marine bacteria produces possibly hundreds of small molecules called secondary metabolites, most of which have never been studied before.
“We’re trying to provide the highest quality microbial extracts from a diverse array of microbes that are producing small molecules that are unusual, different, and that might ‘hit’ the target,” Mincer said.
Hitting the target
The target in this case is a protein called cystic fibrosis transmembrane conductance regulator, or CFTR, which pumps chloride ions out of the cell. In a person with CF, the protein doesn’t pump enough chloride, and the cells produce mucus that is much thicker and stickier than it should be. That clogs up passages, making it harder for sufferers to breathe, for example. The gooey mucus also presents an appetizing environment for infectious bacteria. Repeated, difficult-to-treat infections scar the lungs and are the most common cause of patients’ premature death. Cystic fibrosis afflicts 30,000 Americans and about 70,000 people worldwide.
The disease varies in severity depending on which form of CFTR an individual has. Scientists have identified more than 500 forms of CFTR, each with a different mutation. Mincer and Flatley will focus on the most common mutation, which affects about 70 percent of those with the disease, or 21,000 people in the U.S.
Scientists at Flatley Discovery Lab have developed an assay to test whether a given compound improves the function of this defective form of CFTR. Mincer has started collecting chemicals from bacterial and phytoplankton cultures already on hand at WHOI. Later he will collect new samples from a variety of marine environments, including hydrothermal vents, the open ocean, and salt ponds.
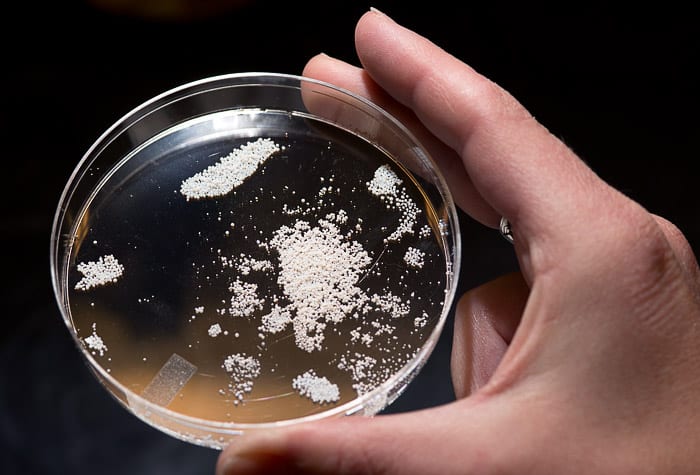
The extraction process is straightforward. The microscopic cells are grown in a nutrient broth that contains a small cloth pouch filled with tiny resin beads. The pouch works like a teabag in reverse: Instead of sending flavorful molecules into the water, the beads soak up the metabolites put out by the microbes. Once a week, technician Kristen Rathjen removes the bags, uses an organic solvent to remove the metabolites, dries the chemicals, and then dissolves them in a carrier solution. That creates a crude extract containing from 300 to 2,000 unique compounds.
Rathjen then runs the crude extract through a High Performance Liquid Chromatography column, which separates the chemicals into 87 groups, or fractions, by their polarity, a measure of their affinity for fats or for water. In most cases, each fraction contains from two to 20 chemicals. The fractions, along with a sample of the crude extract, are dried in a vacuum chamber and sent to FDL for testing. Mincer and Rathjen will also purify compounds that give promising results in the FDL assays and determine their structure, which could provide clues to their function in nature.
Nature’s junk drawer
Mincer said the ability of chemicals from bacteria to affect human cells isn’t surprising, given their shared evolutionary history. A chemical that serves one function in a bacterium may not be needed by higher organisms to do the same function—but the higher organisms may retain the ability to make it or respond to it. Like a piece of string or odd tool stashed in a drawer, eventually it may come in handy.
“Nature does this a lot,” said Mincer. “It’s sort of a parts shop. Stuff gets re-purposed for some new use.”
He plans to use duplicates of the samples he sends to Flatley to study what the metabolites do for microbes in the environment. He’s especially eager to find out whether the bacteria make them all the time, or only when stimulated by specific environmental conditions such as a change in temperature or the presence of predators.
“This is another aspect of the project that I’m really excited about,” he said, “Perturbing the bacteria by saying, ‘Look, here’s something that is going to eat you; what are you going to do now?’ ”
Combining ecology and medicine
This project is not Mincer’s first foray into pharmaceutical discovery. During his graduate work at Scripps Institution of Oceanography he studied marine actinomycetes, a group of bacteria that makes a variety of potent chemicals to ward off grazing organisms such as protists. One day he noticed that a colony of actinomycetes he was growing in the lab was surrounded by dead protists. Realizing that compounds secreted by the bacteria might have been responsible, he passed a bacterial extract along to technicians in the lab who ran it through an anti-cancer screening test. It proved to contain a highly potent killer of reproducing cells.
Mincer and others in the lab isolated the compound responsible (by then called salinosporamide A), patented it, and determined its structure. It is now in phase 2 trials for treatment of multiple myeloma and a rare skin cancer. Along the way, a personal experience brought the significance of the work home to him.
“My in-laws in California have a dear friend who happened to be in the early clinical trials, and the drug has extended his life by several years,” said Mincer. “To see that side of things really influenced me.”
He said that despite such promising results, it’s been difficult for scientists to get funding from federal agencies to do the exploratory work that could lead to further drug discoveries. During his first year at WHOI, Mincer’s requests for support from the National Science Foundation were firmly rejected. “Reviewers said it was a fishing expedition,” he said. “It was hard; I really wanted to do this kind of work. What I really enjoy is being able to bridge that gap between drug discovery and basic research in ecology.”
Support from the Flatley Discovery Lab will enable him to gather samples for his ecological work—perhaps enough of them to interest the large federal agencies—as well as provide ample material for the cystic fibrosis work. The grant extends over three years and will pay for the equipment and full-time technician to extract and fractionate the chemicals. In return, Mincer and his team will send Flatley eight extracts per week—about 700 fractions and thousands of individual compounds—for testing.
That’s a lot of potential drugs, which Mincer said is one of the biggest advantages of using microbes as miniature chemists: the more possibilities you start with, the better your chances of finding a winner. In recent years many large pharmaceutical companies have reduced or eliminated their natural products divisions, he said, choosing instead to invest in efforts to make potential drugs in the lab—but those attempts have struggled to generate enough good prospects to pay their way.
“It turns out, those methods don’t mimic the diversity,” said Mincer. “They don’t even come close to what’s possible from nature.”
This research is supported by a grant from the Flatley Discovery Lab.
Slideshow

Slideshow
- Technician Kristen Rathjen displays flasks in which microscopic marine organisms have been growing for a week. Each flask contains one strain of microbe in a nutrient broth. The broth, which started out pale beige, now appears colored due to the natural pigments of the microbes. (Photo by Tom Kleindinst, Woods Hole Oceanographic Institution)
Each culture flask contains a cloth pouch filled with tiny beads of resin. The pouches act like teabags in reverse, gathering metabolic compounds out of the broth. The microbes growing in this flask were photosynthetic, hence the green pigmentation of the broth.
(Photo by Tom Kleindinst, Woods Hole Oceanographic Institution)Resin beads are tiny but very effective at collecting compounds produced by the microbes. The compounds stick to the beads, which can then be processed to remove the compounds for analysis.
(Photo by Tom Kleindinst, Woods Hole Oceanographic Institution)To remove compounds from the beads, the pouches are placed in an organic solvent, as in the small flasks in the background. The resulting crude extract is then run through a High Performance Liquid Chromatography column (HPLC) to separate the compounds by their polarity. In the foreground, two fresh pouches are ready to be placed in a culture flask.
(Photo by Tom Kleindinst, Woods Hole Oceanographic Institution)Each one-liter culture yields about 3-5 milliliters of crude extract, which is stored in small vials prior to fractionation and analysis. Each extract retains the distinctive color of the culture it came from.
(Photo by Tom Kleindinst, Woods Hole Oceanographic Institution)
Related Articles
Topics
Featured Researchers
See Also
- WHOI Teams with Flatley Discovery Lab WHOI News Release