Down to the Sea on (Gene) Chips
The genomics revolution is transforming the way scientists can study life in the oceans
A half-century ago, James Watson and Francis Crick (aided by Rosalind Franklin and Maurice Wilkins) discovered the double-helical structure of deoxyribonucleic acid (DNA). Other scientists soon showed how DNA—through a triplet code of nucleotide bases on the DNA “spiral staircase” and through ribonucleic acid (RNA) intermediaries—instructs cells to assemble essential life-sustaining proteins. These discoveries opened the door to a new understanding of life by revealing the genetic “blueprints” that underlie the ability of organisms to grow, survive, and reproduce.
A revolution in biotechnology ensued, giving scientists methods to isolate and identify genes, make millions of copies of them, and determine their sequences of nucleotide bases. Together, the accelerating pace of biotechnological advances and the exponential increase in DNA sequence information ignited an explosion in molecular biology and led to the emergence of a new field: genomics. These advances were initially applied in the biomedical arena, leading to new information on the genes responsible for heritable diseases, the molecular signatures of cancer cells, the biology of human pathogens, and genetic factors that influence an individual’s sensitivity to drugs or toxicants.
Now, the genomics revolution has reached the oceans. New genomic techniques are being used to find previously unknown life forms in the oceans; to learn how species, and genes themselves, evolved over Earth’s long history; to understand the genetic tools that allow species to adapt to diverse and often harsh environments; and to investigate species’ responses to pollutants. Genomics gives marine scientists powerful new ways to address age-old questions about life in the oceans.
What is genomics?
Genomics is more than simply determining the sequence of nucleotides in an organism’s genome (the entire set of genetic information contained within a cell’s DNA). It is a new approach to questions in biology, distinguished from traditional approaches by its scale. Rather than studying genes one by one, genomic approaches involve the systematic gathering and analysis of information about multiple genes and their evolution, functions, and complex interactions within networks of genes and proteins.
Genomics has two branches. One is structural genomics—studies of how genes and genomes are organized and how that varies among individuals, populations, and species. It includes characterization of the sequences of DNA nucleotides that encode proteins, as well as the DNA found between and within genes that does not code for proteins.
Using structural genomics, we can compare DNA sequences among individuals of a species to reveal minor variations in the DNA nucleotide code at certain positions in the genome, called “single-nucleotide polymorphisms,” or SNPs (pronounced “snips”). These SNPs can be responsible for genetic diseases, or for hypersensitivity or resistance to drugs or toxicants.
By comparing DNA sequences among species (called “comparative genomics”), scientists can identify changes in genomes that have occurred as species evolved. They can also begin to determine the function of specific DNA sequences shared among different species.
The second branch is functional genomics—the study of the RNA and proteins produced by genes (referred to as “gene expression”), and how these molecules interact to carry out cellular processes.
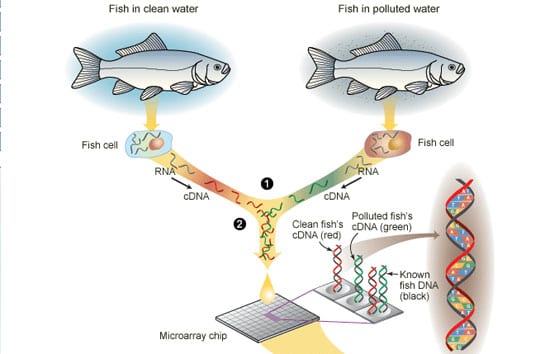
Among the most elegant and widely used tools of functional genomics is the microarray, or “gene chip” (see figure), which became available less than a decade ago. By using microarrays to simultaneously measure the amounts of hundreds or thousands of specific RNAs contained in cells or tissues, biologists can “see” what cells are doing and how they are responding to particular environmental conditions.
Genes reveal marine biodiversity
Though the genomics revolution immediately swept into biomedical research, its entrance into oceanography and marine biology lagged. But marine scientists at WHOI and elsewhere have now begun to take advantage of genomic methods and approaches, aided by three recent developments.
First, the costs of instruments to do genomic research, especially the costs of DNA sequencing, have declined dramatically over the past five years. Second, efforts to sequence genomes have started to include more marine species, from bacteria to animals. And third, recent studies have shown that genomic approaches can be used even if a species’ genome sequence is not yet known. Marine organisms with unusual or unique adaptations, for example, can now be studied using genomics. In addition, by sequencing DNA from samples of seawater or ocean sediments, scientists can find new organisms by comparing the newfound genes with similar gene sequences of known organisms.
Scientists are hampered in the study of single-cell microbes, because it is often difficult to replicate the microbes’ undersea conditions in the laboratory and culture the microbes successfully. But molecular and genomic approaches are yielding important and sometimes surprising information about the diversity, abundance, and ecological roles of marine microbes in diverse environments, including extreme environments such as the deep sea and polar regions. (See “The Deeps of Time in the Depths of the Ocean”.)
Recently, the genomes of several marine microbes have been sequenced, providing a window on their genetic, biochemical, and physiological adaptations to their diverse physical, chemical, and biological environments. Some of the microbes’ unusual physiological abilities, inferred initially from genomic sequences taken from environmental samples, have been confirmed subsequently by detailed study of expressed proteins.
For example, Ed DeLong’s research team at the Monterey Bay Aquarium Research Institute isolated genes encoding a novel type of light-harvesting pigment that mediates an unusual form of photosynthesis in marine bacteria. Genomic studies of uncultured bacterial samples from oceanic waters showed that these pigments are much more diverse and widespread than expected. These findings are changing the way we think about the importance of marine bacteria in the flow of carbon and energy in marine ecosystems, including their possible role in the uptake of atmospheric carbon dioxide.
Genomic insights into evolution
Genomic information also provides a key to unlocking longstanding mysteries of evolutionary history. Scientists are comparing the characteristics of genomes—DNA sequences, gene structure, and chromosomal organization—to infer evolutionary relationships among organisms. Such studies are helping scientists construct the tree of life, which traces the evolution of organisms from ancestral single-celled beginnings to the diversity that exists today.
Such studies have shown that an organism’s DNA can come not just from its direct ancestors but also from distantly related species. Though genes are transferred vertically (from parent to offspring), a surprising 17 percent of the DNA in some bacterial species has been acquired by horizontal gene transfer (between species). Genes encoding bacterial antibiotic resistance, a major problem in hospitals, are known to be passed around in this way, but the implications of horizontal gene transfer in the marine environment are not yet well understood.
To understand age-old evolutionary mysteries of how animals evolved such a variety of body plans—from jellyfish to mollusks, crustaceans, worms, and vertebrates—biologists are now using genomics to elucidate the structure, expression, and evolutionary history of the genes responsible for generating morphological diversity during embryonic development. Genomic studies in a variety of animals, including tunicates (marine invertebrates that are close relatives of vertebrates), have helped reveal the importance of a set of genes known as the Hox cluster. The specific positions of the Hox genes on the chromosome influence how these genes are expressed during embryonic development and ultimately how they affect the shape of the embryo.
Genome sequencing in animals, plants, and fungi also has revealed that at certain times in evolutionary history, various lineages developed duplicate genomes—that is, extra sets of the same genes. The extra genes increase the chance that mutations providing advantageous anatomical, physiological or biochemical traits will be retained. In this way, genome duplications may promote biodiversity.
Genomic studies have shown that one or two whole genome duplications occurring more than 450 million years ago may have facilitated the evolution of vertebrates (animals with backbones). Another more recent genome duplication may have led to the extraordinary diversity of bony fishes (about 30,000 species), which account for about half of vertebrate species.
Genomic clues to symbiosis
Genomics is also providing insights into the factors that underlie symbiosis, the fascinating interrelationships between two organisms that can be either mutually beneficial (mutualism) or harmful to one member of the pair (parasitism). Genomic sequencing of symbiotic organisms has shown that dramatic changes in genome structure often occur during the evolution of their association. Parasites, for example, often exhibit reductions in genome size and gene number, accompanied by the loss of the ability to carry out certain biochemical reactions.
Scientists are also using microarray-based measures of gene expression to understand symbiotic mechanisms between reef-building corals and dinoflagellates that help corals grow and build their skeletons. The studies will also yield better understanding of coral bleaching, which occurs when the dinoflagellates leave or are ejected from the corals.
Adapting to the environment
Scientists have learned that changes in an organism’s environment, including pollution, often elicit compensatory changes in the expression of specific genes. For example, Andrew Gracey and George Somero at Stanford University’s Hopkins Marine Station have used microarrays to identify genes involved in the ability of a burrow-dwelling fish, the goby (Gillichthys mirabilis), to adapt to the reduced oxygen levels (hypoxia) that occur in its intertidal burrows.
Similarly, scientists at WHOI are studying the genes and proteins involved in the response of fish to contaminants such as chlorinated dioxins and polychlorinated biphenyls (PCBs). At WHOI, Heather Handley-Goldstone and John Stegeman have used microarrays to identify genes associated with heart malformations caused by dioxin in embryonic zebrafish.
In my lab, we use genomic techniques to trace the evolution of genes encoding receptor proteins through which dioxins and PCBs cause altered gene expression and toxicity. We have found that fishes have more of these receptor genes than mammals, possibly explaining the extreme sensitivity of fish to these chemicals. At the same time, we are also studying how small changes in these genes—SNPs—might be involved in PCB resistance that sometimes develops in Atlantic killifish (Fundulus heteroclitus) living in highly contaminated sites such as the harbor in New Bedford, Mass.
From genomics to bioinformatics
Despite the progress in applying genomics to marine systems, challenges remain. One of the most significant is dealing with the huge amounts of data generated in genomic experiments, which must be analyzed in relation to environmental or physiological measurements collected at the same time. To accomplish this, we use bioinformatics—mathematical and computational methods for analyzing and visualizing genomic data.
As bioinformatic approaches become more sophisticated and essential, biologists without extensive mathematical backgrounds will need enhanced training in computational genomics and bioinformatics. We will also need to recruit more mathematicians and computer scientists into the marine science community.
The conceptual and technical advances associated with genomics are revolutionizing research in biology and medicine. The emergence of genomics gives the marine science community an unprecedented opportunity to address old questions in new ways and to formulate new questions stimulated by our expanding genomic understanding of life in the oceans.
Related Articles
Topics
See Also
- National Institutes of Health Genomic Biology
- Ocean Genomics from Nature magazine
- Test for Dioxin Sensitivity in Wildlife Could Result from New Study WHOI media release
- Ocean Life Institute
