
Symbiosis in the Deep Sea
Scientists discover how bacteria living with shrimp make a living
Mobs of pale shrimp clamber over each other, jockeying for position in the swirling flow of black-smoker vents on the seafloor where ultra-hot fluids from Earth’s interior meet cold seawater.
The shrimp, Rimicaris exoculata, are not the only animals that live near these vents on the Mid-Atlantic Ridge, the volcanic seafloor mountain range that bisects the ocean basin. But they are by far the most abundant, with population densities reaching 3,000 individuals per square meter. Though numerous and known to science since 1986, how they make their living is still a mystery.
At first, scientists thought they grazed on bacteria that grow on the chimney-like walls of the vents, which spew dark, chemical-rich fluids into the ocean. Then, in the 1990s, a novel kind of bacterium was found living in the shrimps’ gill chambers. That finding raised the possibility that Rimicaris relies on symbiotic relationships with bacteria, which, in turn, need chemicals in the vent fluids to live.
But figuring out how two organisms interact requires more detailed knowledge than just seeing them together. What kinds of bacteria are present with the shrimp? What, if anything, do the bacteria and the shrimp provide to each other? How do they do it?
Stefan Sievert, a microbiologist at Woods Hole Oceanographic Institution (WHOI), and colleagues in Europe recently took a big step toward answering those questions about Rimicaris. They found that the shrimp’s mouths and gill chambers harbor a more diverse community of bacteria than previously thought. These bacteria, called epibionts, have a wide range of metabolic abilities that could supply the shrimp with organic carbon and other nutrients.
The project came about when Mike Atkins, an oceanographic consultant and graduate of the MIT/WHOI Joint Program in Oceanography, joined film director James Cameron’s cruise to the “Snake Pit” vent field during the making of the documentary Aliens of the Deep.
“Mike contacted me and asked if we would like to have some samples,” said Sievert, who usually studies free-living microbes in vent ecosystems. “I was always fascinated by these shrimp, so I thought maybe we could do something with the epibionts. That’s how it started—getting in these samples by chance, more or less.”
Clues from genes
Atkins sent Sievert several shrimp that had been frozen to preserve their DNA as soon as they were brought on the ship. Sievert said these shrimp look much like the ones that end up in shrimp cocktails. “But not as tasty! They are coming from a very sulfidic place, and you can already smell it when you thaw out the samples.”
The first step in determining whether bacteria and shrimp interacted or were together just by chance was to find out what kinds of bacteria were present and where. Sievert and Michael Hügler, then a postdoctoral scientist in Sievert’s lab, dissected the gill chambers of four shrimp, separating their two main structures, the mouth parts and the inner lining of the chamber wall. If bacteria were randomly distributed on the parts, they were probably just passing through; if specific kinds of bacteria were associated with specific areas, there might well be a biological reason.
Then Hügler extracted DNA from the bacteria to identify what kinds of microbes were present. He and Sievert found at least ten different kinds of bacteria from four major taxonomic groups living in the shrimps’ gill chambers. One kind was found almost exclusively on the inner lining, one was found almost exclusively on the mouth parts, and the others were found, in varying proportions, in both locations.
Next, they looked for whether each kind of microbe had genes that code for enzymes needed in specific biochemical reactions to metabolize carbon, sulfur, and nitrogen. Several biochemical pathways can be used to oxidize sulfur, for instance, and each one can help reveal how energy and nutrients are used by various vent organisms.
They found that the bacteria living in the gill chambers employ at least three mechanisms for gaining energy. Some do it by oxidizing compounds. Some oxidize molecular hydrogen, others oxidize sulfur compounds that have been reduced (the opposite of oxidized), and some might do both simultaneously. At least one type of bacterium makes a living by reducing oxidized sulfur compounds. That raises the possibility that two or more kinds of bacteria engage in a localized “sulfur cycle”—working efficiently together by providing each other with essential substrates and in the process increasing the amount of biomass they can produce. They also found evidence that bacteria use two different pathways for converting carbon dioxide into organic compounds.
A different perspective
A few months into the molecular work, Sievert’s lab had a roster of the bacterial community, good clues about their metabolisms, and a general sketch of where various kinds of bacteria were in the shrimp. Then he crossed paths with colleague Nicole Dubilier from the Max Planck Institute for Marine Microbiology in Bremen, Germany, who came to WHOI for a conference and gave him a way to actually be able seethe microbes.
Sievert knew Dubilier while in graduate school at the Max Planck Institute and the two had worked together several times over the years. When Sievert mentioned his shrimp project, Dubilier said that Christian Borowski from her lab had also been on Cameron’s cruise and had preserved shrimp in a different way to do a technique called Fluorescence In Situ Hybridization, or FISH.
“We didn’t have the kind of samples that they had, and they didn’t have the kind of samples that we had, so we said, why don’t we bring them together and make a more convincing story?” recalled Sievert. “It’s interesting how science works sometimes. You try to be very careful and plan everything, but sometimes it happens by coincidence and sheer luck.”
FISH links a fluorescent tag to specific DNA or RNA probes that will recognize and bind to their unique corresponding gene sequences in target cells on a microscope slide. In this case, the targets were the bacteria in thin slices of shrimp tissue. Viewed through a special microscope, the tag lights up and reveals the location of that particular sequence. Researchers can use probes carrying different-colored tags to visualize the locations of more than one sequence at a time.
The FISH studies were carried out by Jillian Petersen, at the time a Ph.D. student in Dubilier’s lab. The images showed long strands of brightly labeled bacteria attached in clusters to the shrimp’s mouthparts, each kind of microbe in a specific location. The view through the microscope confirmed what the molecular work in Sievert’s lab had suggested: Many or most of the kinds of bacteria he found in the gill chambers are not there by accident. They probably serve a critical function for the shrimp.
Sievert said he thinks the shrimp and their bacterial hangers-on probably have the kind of symbiotic relationship known as mutualism, in which both participants benefit. It’s possible, for instance, that the shrimp get organic matter—food—from the bacteria, either eating them directly or consuming their waste products; the shrimp could act as platforms that keep the bacteria in prime locations, where temperatures and geochemical conditions are just right.
Sievert plans to go back to the deep-sea vents on the Mid-Atlantic Ridge to test some hypotheses that resulted from this work and to look more directly at the activity of the microbes involved. Sievert is also leading an interdisciplinary and multinational team of investigators funded by the Dimensions of Biodiversity program of the National Science Foundation to learn more about how deep-sea vent microbial communities function and how they affect the larger ocean.
This research was supported by the U.S. National Science Foundation and by the Alfried Krupp Wissenschaftskolleg, the Max Planck Society, the German Research Foundation, MOMARnet, and IFM-GEOMAR in Germany.
Slideshow
Slideshow
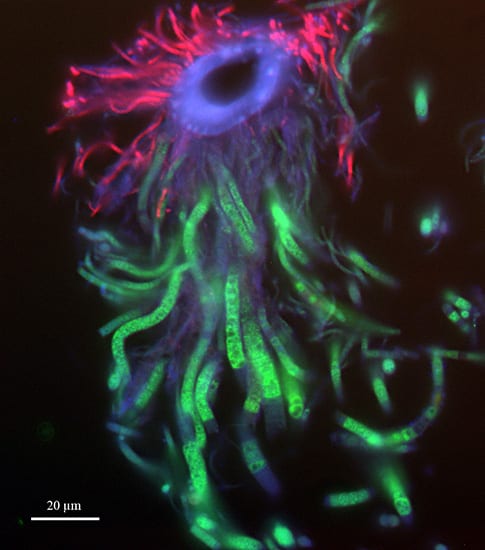
 WHOI microbiologist Stefan Sievert and colleagues used FISH, or Fluorescence In Situ Hybridization, to localize and identify the microbes living in different parts of the gill chambers of shrimp living near hydrothermal vents. In the FISH technique, short stretches of nucleic acids tagged with different fluorescent labels bind to the DNA of specific microbes. This image shows two kinds of bacteria attached to a hair-like structure called a seta on the mouth appendages of the shrimp Rimicaris exoculata. The ring-shaped seta (seen here in cross-section) appears blue. Short, thin Gammaproteobacteria appear red, and longer, stouter Epsilonproteobacteria appear green. The scientists are working to determine whether these bacteria convert energy from chemicals in hydrothermal fluids into biomass that the shrimp can use as an energy source. (Image by Jillian Petersen, Symbiosis Group, Max-Planck-Institute for Marine Microbiology, Bremen, Germany)
WHOI microbiologist Stefan Sievert and colleagues used FISH, or Fluorescence In Situ Hybridization, to localize and identify the microbes living in different parts of the gill chambers of shrimp living near hydrothermal vents. In the FISH technique, short stretches of nucleic acids tagged with different fluorescent labels bind to the DNA of specific microbes. This image shows two kinds of bacteria attached to a hair-like structure called a seta on the mouth appendages of the shrimp Rimicaris exoculata. The ring-shaped seta (seen here in cross-section) appears blue. Short, thin Gammaproteobacteria appear red, and longer, stouter Epsilonproteobacteria appear green. The scientists are working to determine whether these bacteria convert energy from chemicals in hydrothermal fluids into biomass that the shrimp can use as an energy source. (Image by Jillian Petersen, Symbiosis Group, Max-Planck-Institute for Marine Microbiology, Bremen, Germany)- Stefan Sievert, a microbial ecologist at Woods Hole Oceanographic Institution, usually studies free-living bacteria at hydrothermal vent fields. He recently began working on bacteria that live inside shrimp and that may have a mutually beneficial symbiotic relationship with the shrimp. (Photo courtesy of Stefan Sievert, Woods Hole Oceanographic Institution)
Related Articles
Featured Researchers
See Also
- Stefan Sievert's lab
- WHOI Ocean Topics: Hydrothermal Vents
- Nicole Dubilier
- Max Planck Institute for Marine Microbiology
- The journal article "Patterns of Carbon and Energy Metabolism of the Epibiotic Community Associated With the Deep-Sea Hydrothermal Vent Shrimp Rimicaris exoculata"
- Dimensions of Biodiversity PDF describing new research being led by Stefan Sievert as part of NSF's biodiversity program

